A couple of years ago, we started exploring how we could modernise the way NMR data is stored, managed, and accessed at the Imperial College Chemistry Department NMR facility. Back then, things were still very much in prototype mode — we were asking: Is this the right platform? Does it fit the way our facility operates? That early exploration led us to adopt NOMAD, a web-based system designed for storing and sharing NMR data, developed by Tomas Lebl at the University of St Andrews.
In this post, we’ll give a quick overview of why we chose NOMAD and what it’s helping us achieve, then take a peek behind the curtain at the often invisible but very real work needed to make a system like this reliable for hundreds of users.
Why NOMAD?
Before NOMAD, our data management relied on local storage and shared folders — a model that works fine at small scale but quickly becomes messy when hundreds of researchers rely on the facility. It’s difficult to keep track of what’s where, who has access, or even whether data will still be available in a year’s time.
NOMAD provides a web portal for structured data storage, where each experiment is automatically saved with its associated metadata (things like instrument, date, experiment type, and sample ID). That means the data adheres to FAIR data principles — a set of widely adopted standards for making research data better structured, more open, and more useful.
We’re also starting to build an API layer on top of NOMAD, which opens the door to programmatic access. That’s a big deal for anyone working on machine learning — data can be pulled straight into analysis pipelines rather than downloaded and wrangled manually.
Further reading: NOMAD platform homepage
Headline Numbers
Since rolling NOMAD out to the NMR facility, usage has grown rapidly.
| Metric | Value (as of October 2025) |
|---|---|
| Number of registered users | 340 |
| Total experiments stored | 23,600 |
| Average experiments recorded per week | 970 |
| Total data volume | 42 GB |
The scale of these numbers underline why having a proper data management system matters. But it also highlights another reality: to make a system like this reliable, you need a lot more than just the software.
Making It “Production Ready” (aka All the Unseen Work)
When we first set NOMAD up, it was just enough to get the data in and the portal working. But to make it something we — and the entire department — can rely on day-to-day, we had to tackle a lot of extra work behind the scenes.
1. Modernising the Facility
Our spectrometer control PCs were running on operating systems that pre-dated the NOMAD platform itself. To get them onto the college network — a prerequisite for connecting to the NOMAD server — we had to bring their OS and NMR software up to date, replacing unsupported systems and updating software that hadn’t been touched in years.
Why does this matter? Outdated systems create what software engineers call technical debt — essentially a backlog of old software, fragile setups, and workarounds that make it harder to build new things
💬 Explainer: What is “Technical Debt”?
Think of technical debt like scientific instrument maintenance.
If you put off cleaning the NMR probe or recalibrating the spectrometer, experiments still run — for a while — but problems quietly accumulate.
In software, “technical debt” is the cost of postponing updates, refactors, or documentation. Over time, outdated systems, fragile dependencies, and workarounds start to slow down new development — just like an instrument that’s overdue for service.
Why it builds up faster in academia
- Research software often evolves around grant cycles, not product lifecycles.
- There’s limited dedicated engineering time.
- Systems that “still work” can stay untouched for years.
Paying off this debt — by modernising operating systems, updating dependencies, and documenting workflows — keeps the infrastructure reliable and future-proof.
With the modern tech in place, we were able to make the platform much more resilient. Systems now recover automatically from common errors, updates can be rolled out without major disruptions, and compatibility issues that once slowed us down are dramatically reduced. This kind of foundation isn’t just nicer to work with — it’s what makes long-term reliability possible.
2. Backups: Protecting Thousands of Hours of Work
When your data is critical to research, backups aren’t optional. We put in place a multi-layer backup strategy to make sure nothing is lost:
- Daily snapshots of the NOMAD database and file storage.
- Offsite replication so a single server failure isn’t catastrophic.
Automated retention schedules to keep recent backups handy and archive older ones.
More details: Server Backups documentation
3. Monitoring and Alerting: Finding Problems Before They Find Us
A system serving hundreds of users can’t rely on someone noticing “something looks weird.” We set up a monitoring stack based on Telegraf, InfluxDB, and Grafana (TIG stack).
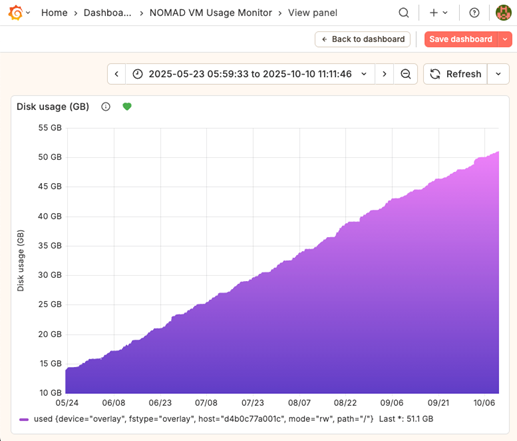
This gives us real-time insight into:
- Server health and uptime
- Storage usage (including that growing data volume chart above!)
- Error logs and system events
And if something does go wrong, alerts are sent directly to our Slack channel, so the right people know immediately.
More details: TIG Stack monitoring documentation
Pulling It All Together
The diagram below shows how all these components fit together — from NMR spectrometers to the NOMAD database, backups, and monitoring.
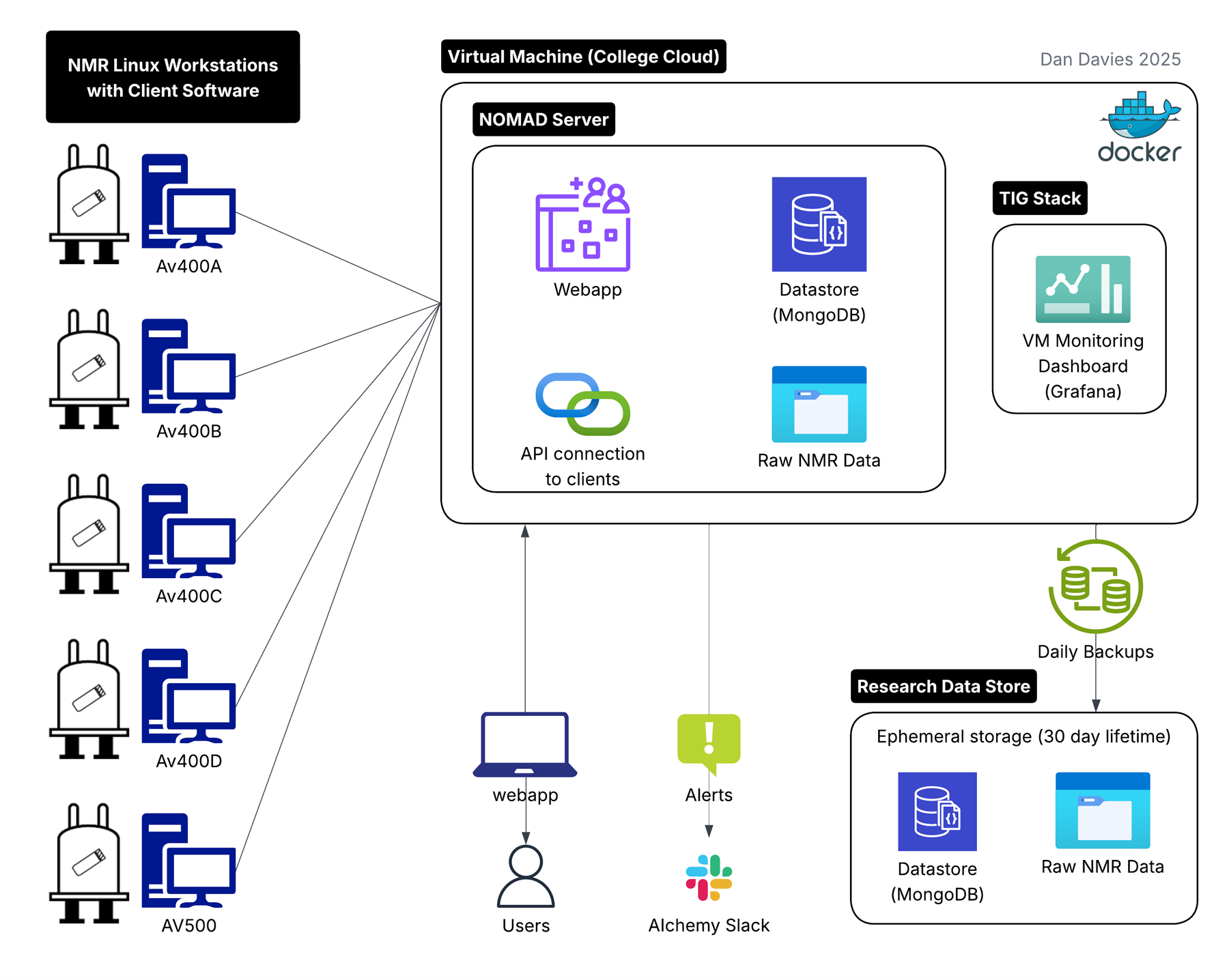
What started as an experiment to see whether NOMAD might work for us has grown into a fully operational, monitored, backed-up, production-grade system supporting hundreds of researchers and thousands of experiments.
Adopting good software is only step one. The real work often lies in everything that surrounds it — upgrading infrastructure, securing data, setting up reliable monitoring, and reducing technical debt so we can keep building in the future.
Written By:


